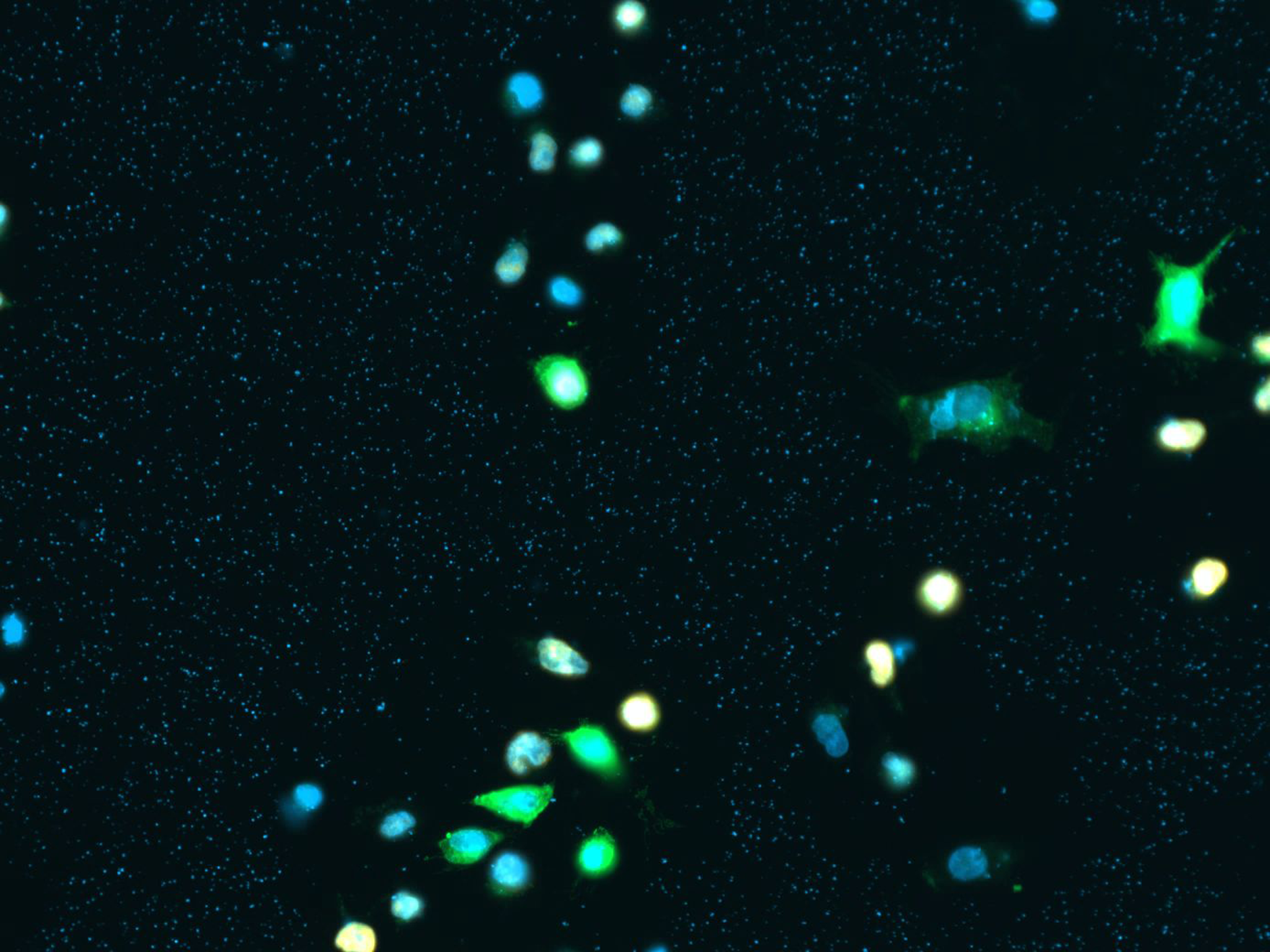
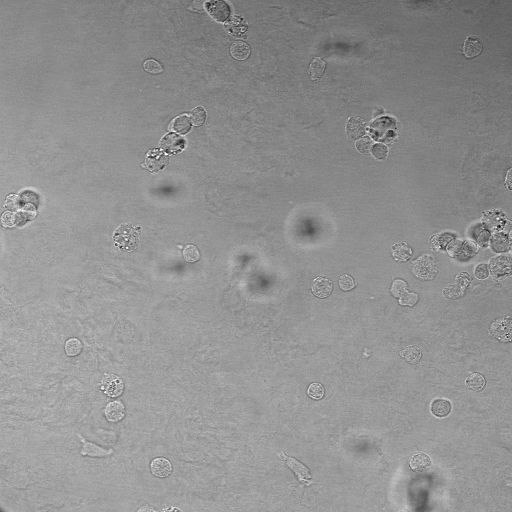
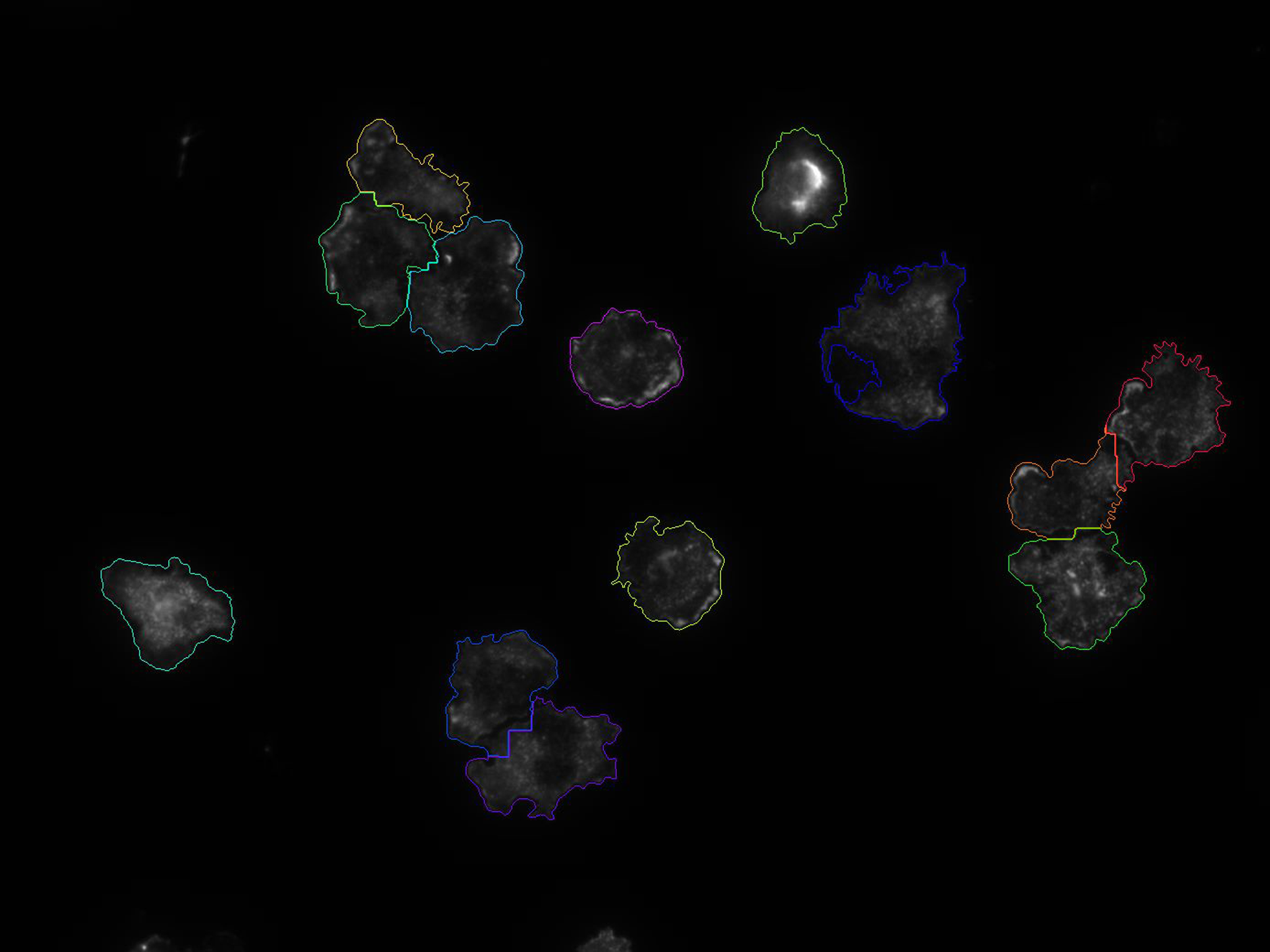
Proliferation / Colocalization
Problem: manually counting cells that express multiple fluorescent markers is tedious work and prone to errors and intra/inter observer variance
Solution: automatically analyze scans and create Excel file
Current Status:
- Successfully employed in practice
- Unterer B, et al. IFN--response mediator GBP-1 represses human cell proliferation by inhibiting the Hippo signaling transcription factor TEAD. Biochem J. 2018 Aug 17
- Prototypical standalone software available
- C/C++ API in development
Solution:
- Large modular toolbox with ~50 image processing building blocks available. We combine and tune these building blocks in order to create powerful image processing algorithms that can be stored as presets/templates
- The solution is free of AI and so does not require a large annotated ground truth database
- Optionally, an image processing pipeline can be automatically optimized by providing ground truth annotations for one or a few sample images