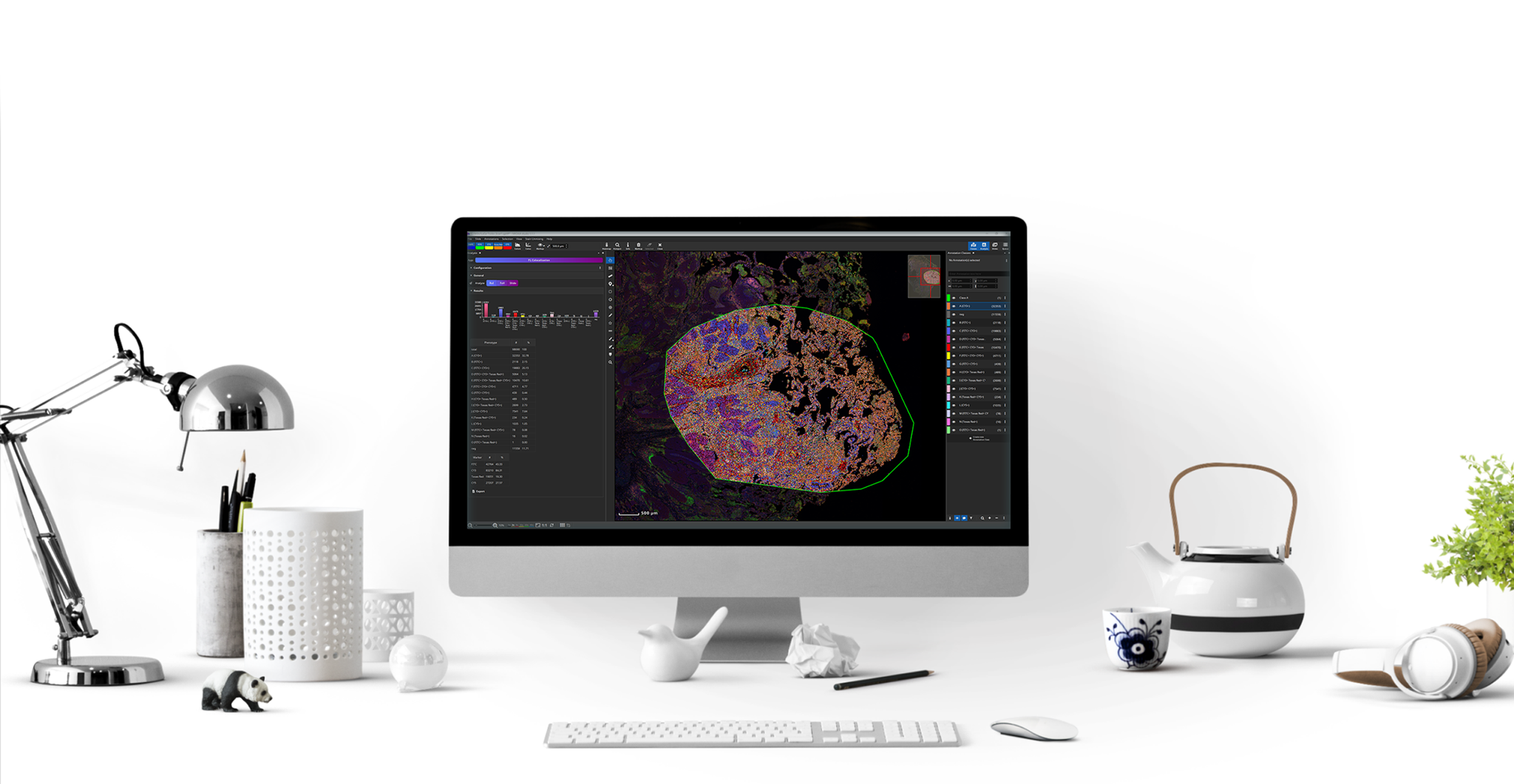
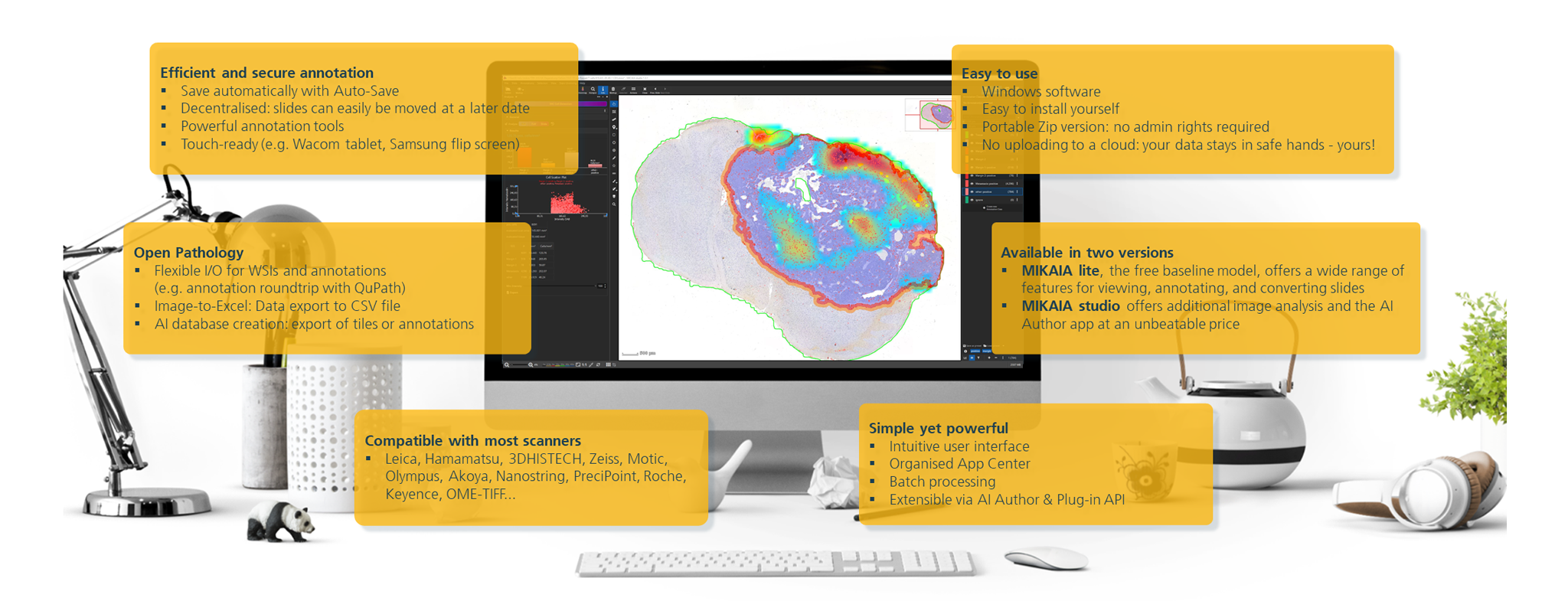
MIKAIA1 is the ideal software for preclinical and clinical research – precisely tailored to the requirements and specific research questions of histopathology:
- Annotation of whole slide images, in brightfield or fluorescence.
- Efficient analysis of more data – for objective and quantitative results.
- Image-to-Excel functionality for quantitative pathology.
- "Off the Shelf" apps for standard use cases as well as flexible extension options for individual use cases.
- Ideal for standard pathology (H&E), immunohistochemistry, and proteomics/patial biology (mIF).
Application notes and use cases are available from the central learning hub MIKAIA University and more resources in our life science blog. The underlying AI technologies are described in our scientific publications.