The biggest challenge in pathology is the shortage of skilled workers while the workload is continuing to grow. New treatment options, particularly in cancer therapy, are based on complex diagnostic procedures that demand an increased number of biopsies and biomarker assessments.
AI-based image analysis in digital pathology
Artificial intelligence and image analysis software play a crucial role in supporting pathological diagnosis and research by offering
- simplified data management: easier archiving, sharing, and accessing of data through the digitalization of specimens
- faster diagnosis and better quality assurance through telepathology
- improved visualization of tissue samples, assistance in their classification, analysis, and interpretation
- automated diagnostic tasks
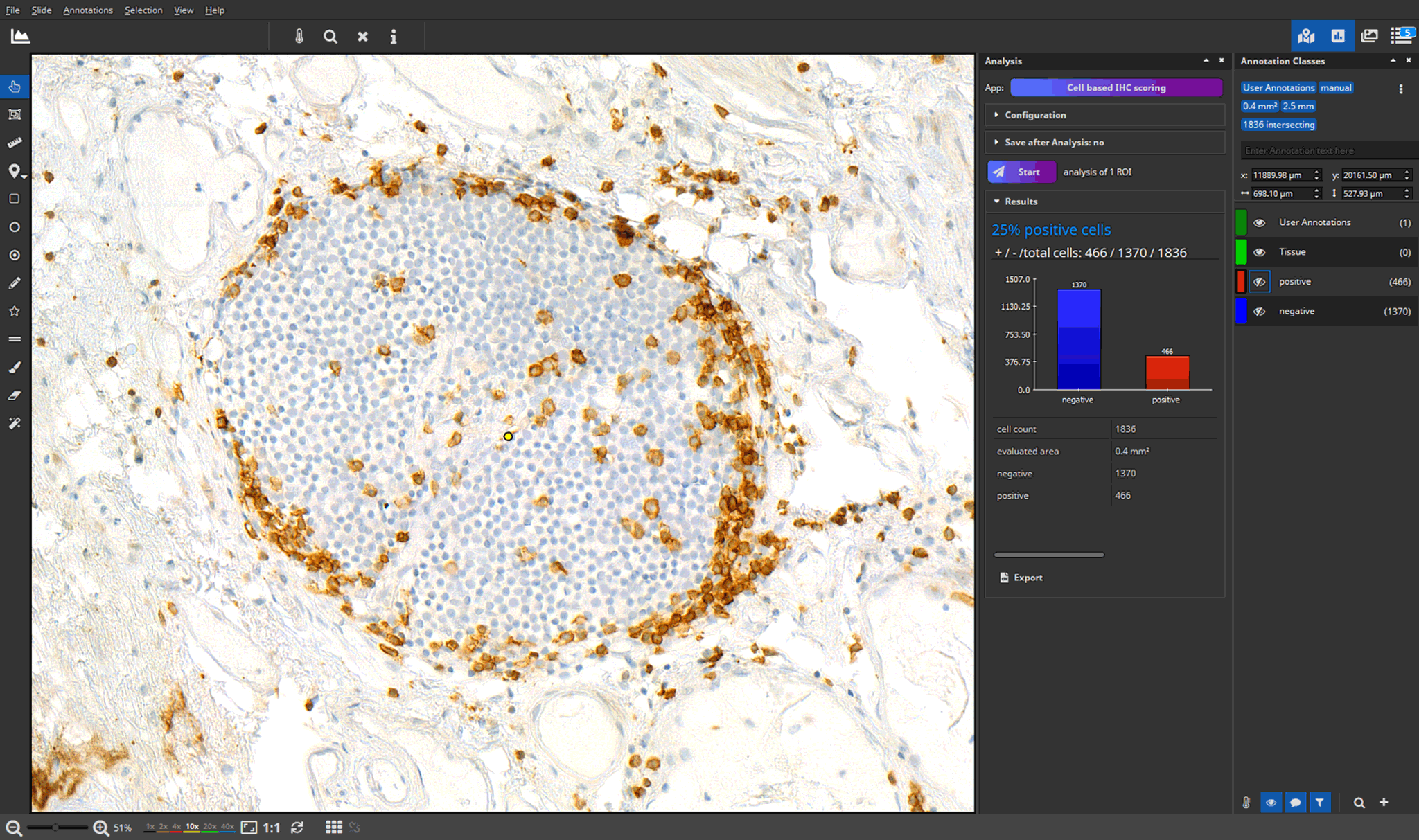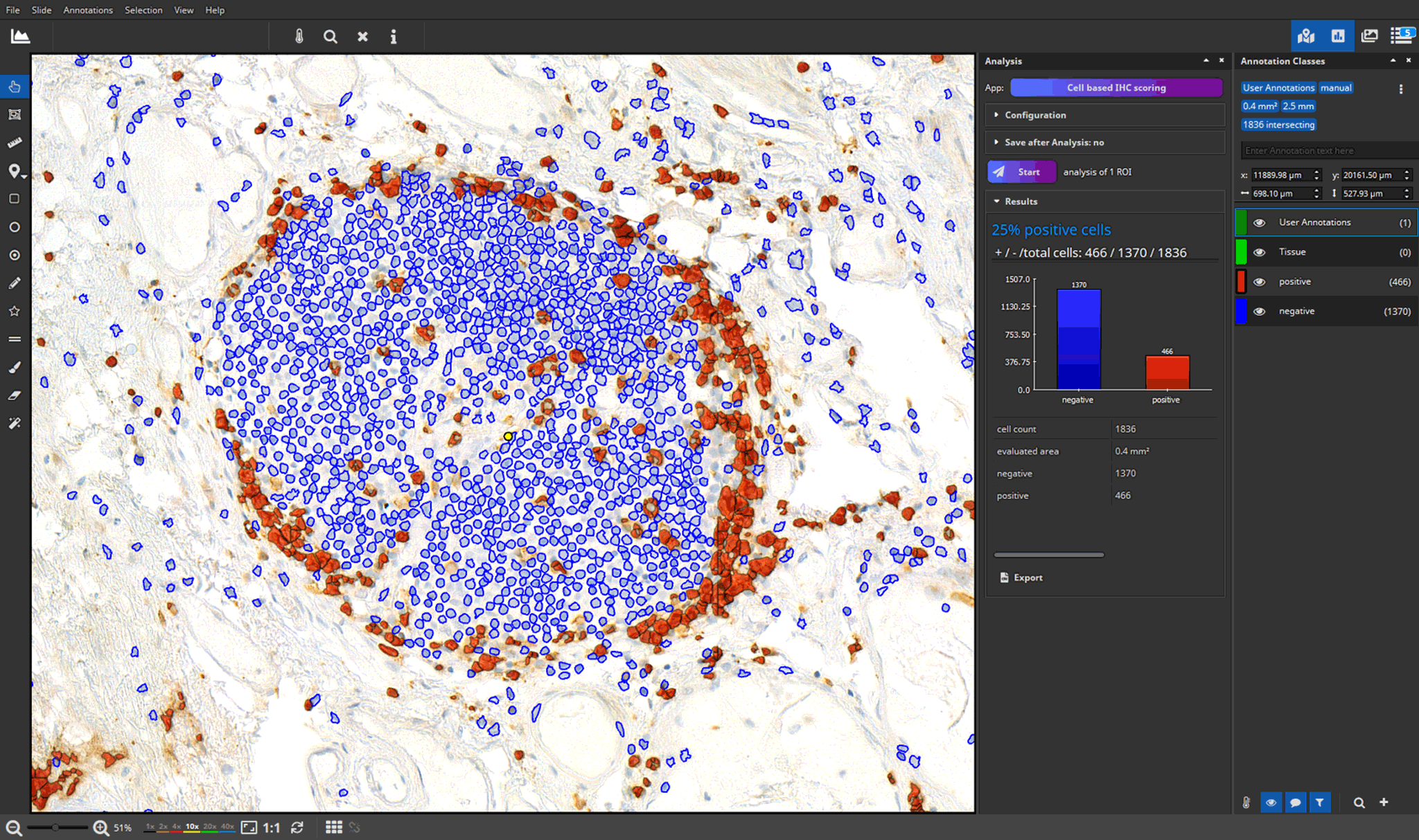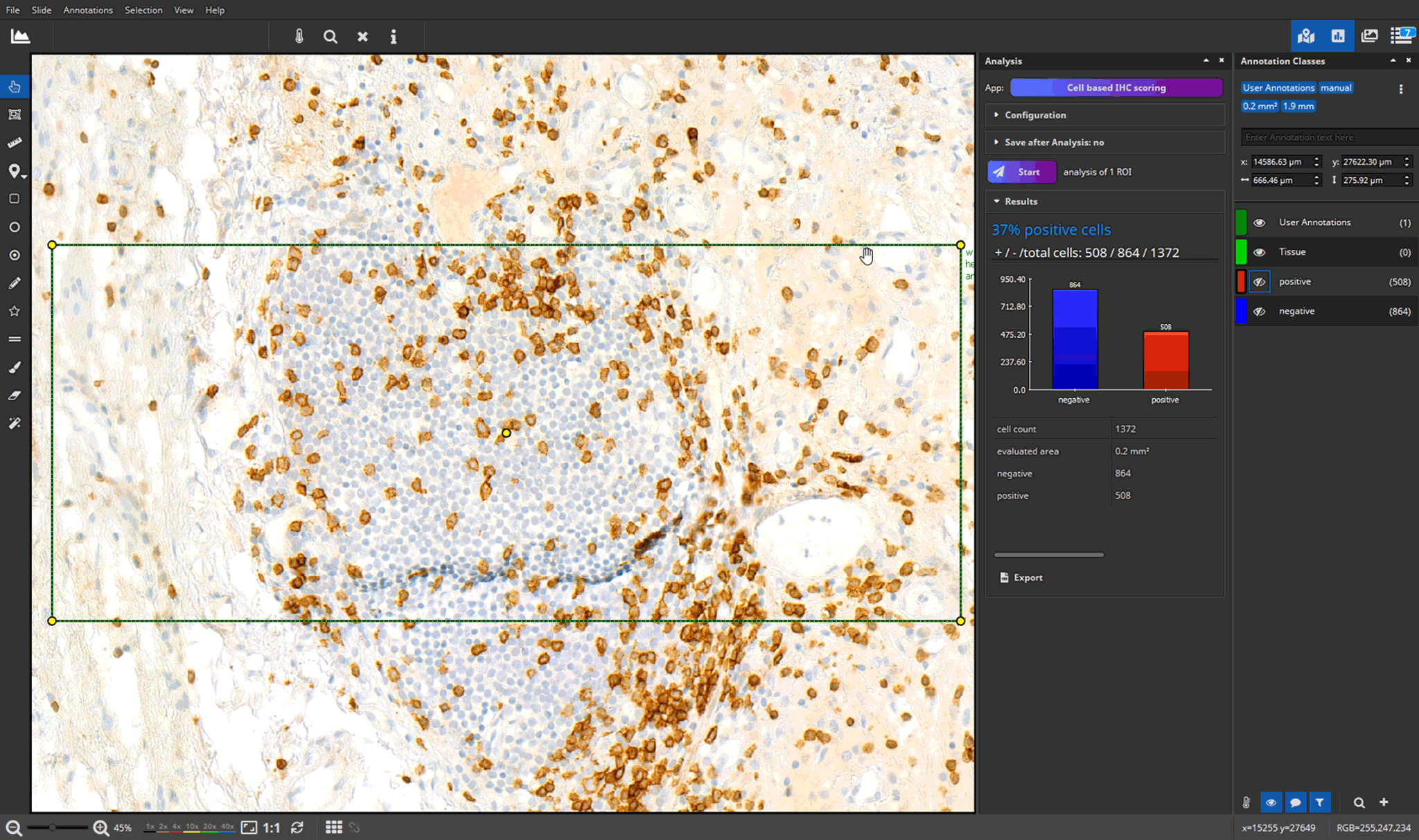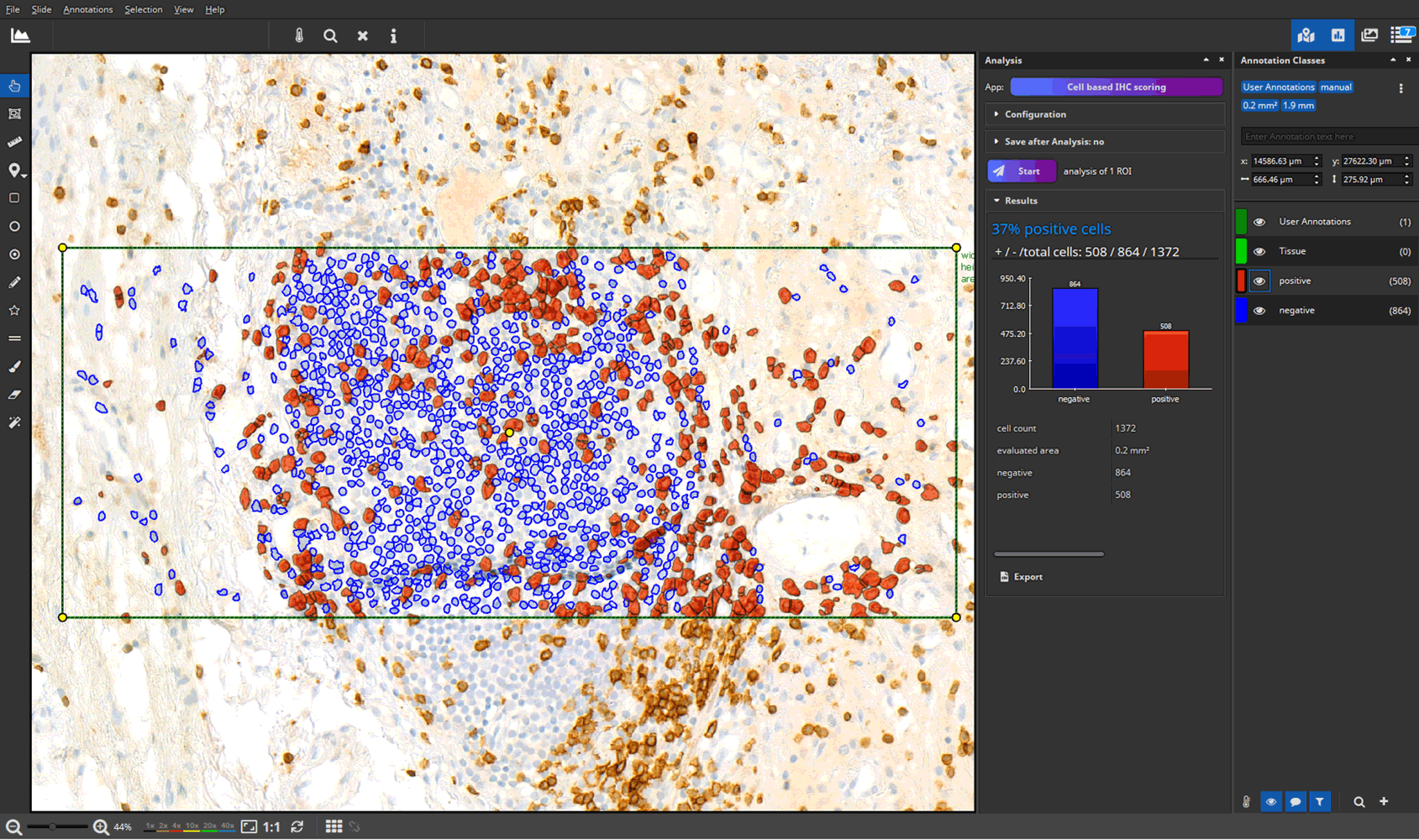
- efficient annotation and real-time visualization of quantitative analyses
- faster and more accurate analysis results through AI-powered pattern recognition
- Spatial Biology techniques: quantitative analyses of the spatial organization and dynamics of biological systems
Medical Image Analysis – Our Offer
We offer fixed-price project development, software and technology licensing as well as analysis and consulting as a service.